15+ pymol coloring
Web I was wondering if there is a way to colour by a structure by hydrophobicity. Web You can change the b-factors column and put your values and then in Pymol use the option Color by B-factor.
Pyproga A Pymol Plugin For Protein Residue Network Analysis Plos One
The convenient way is using the spectrum function to have rainbow.
. First define a set of colors using your features remember to normalize your feature values to scale. Begin a new PyMOL session and open alphafold_coloringpy by clicking File--open in the PyMOL menu bar. Selection-expression or name-pattern corresponding to the atoms or objects to be colored default.
Web Most new PyMOL users like to color the proteinDNAcomplex object with multiple colors. Web 1 Answer. Web By default this will run the script on all atoms in the PyMOL session.
Web color string. Color name or number. You will have then a gradient of colors where the warmer the.
Please refer to pymolwiki page Color and Set Color. Hi Carly This is a quick hack I wrote some time ago but it kind of does the job.

Five Cool Features In Pymol That You May Have Missed Michael S Bioinformatics Blog

Pymol Assign Secondary Structural Regions My Software Notes
Pymol Tutorial Hydrophobicity
Alphafold2 Github Topics Github

How Can I Highlight Some Residue In Stick Model On The Surface Of Protein In Pymol Researchgate

Simple Tools For Mastering Color In Scientific Figures The Molecular Ecologist
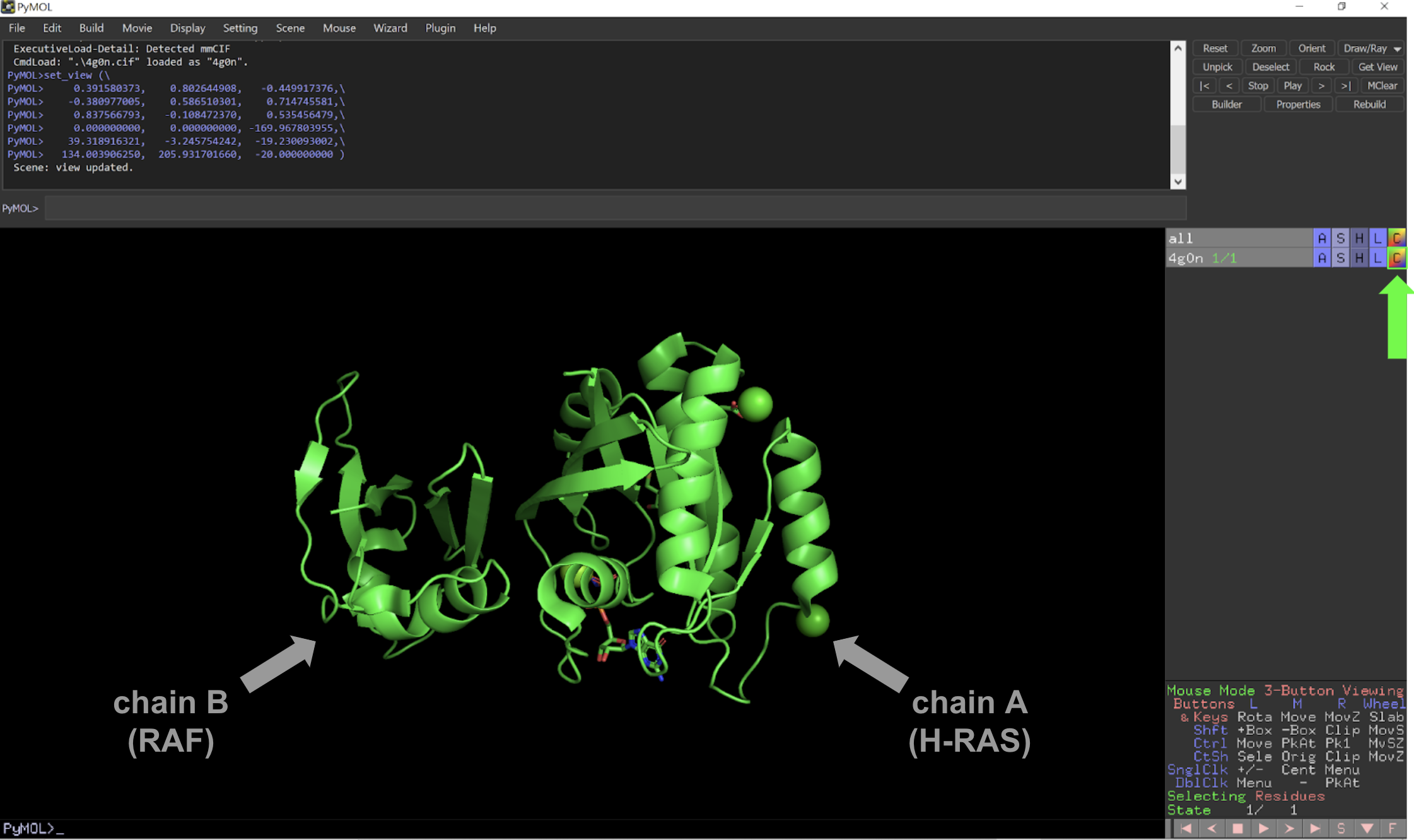
Analyzing A 3d Structure File Labxchange

Media Surfacecolorsmoothing Pymol Documentation
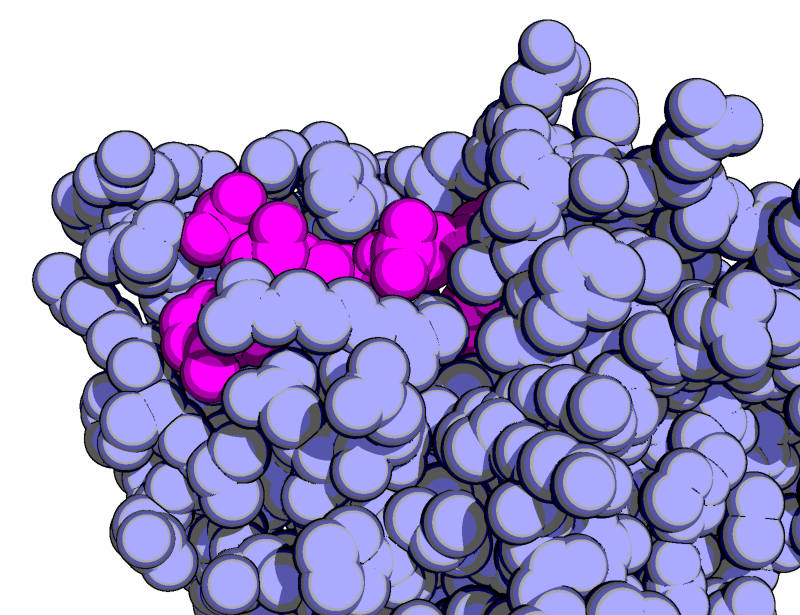
Gallery Pymol Wiki

Pymol Customize Spectrum Colors My Software Notes
Pymol Quick Howto
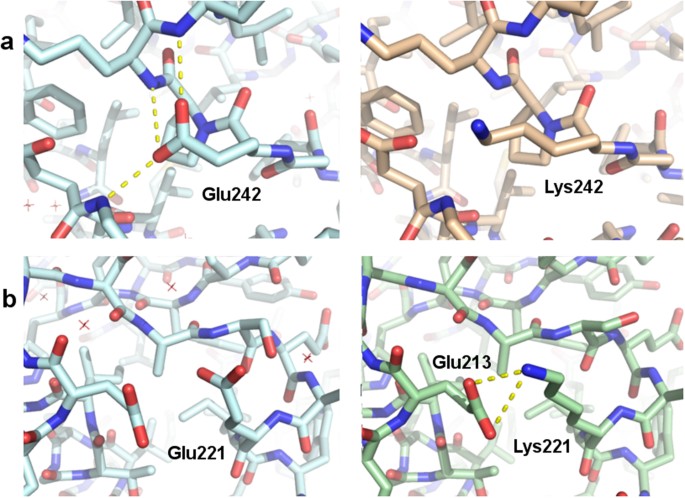
Consequences Of Point Mutations In Melanoma Associated Antigen 4 Mage A4 Protein Insights From Structural And Biophysical Studies Scientific Reports

Coloring Molecule With Resp Charge On Pymol Pymol Is Life Worth Living

Pymol Molmol Like Colors On Secondary Structural Elements My Software Notes

Making Pretty Pictures With Pymol Oxford Protein Informatics Group
Pymol Tutorial
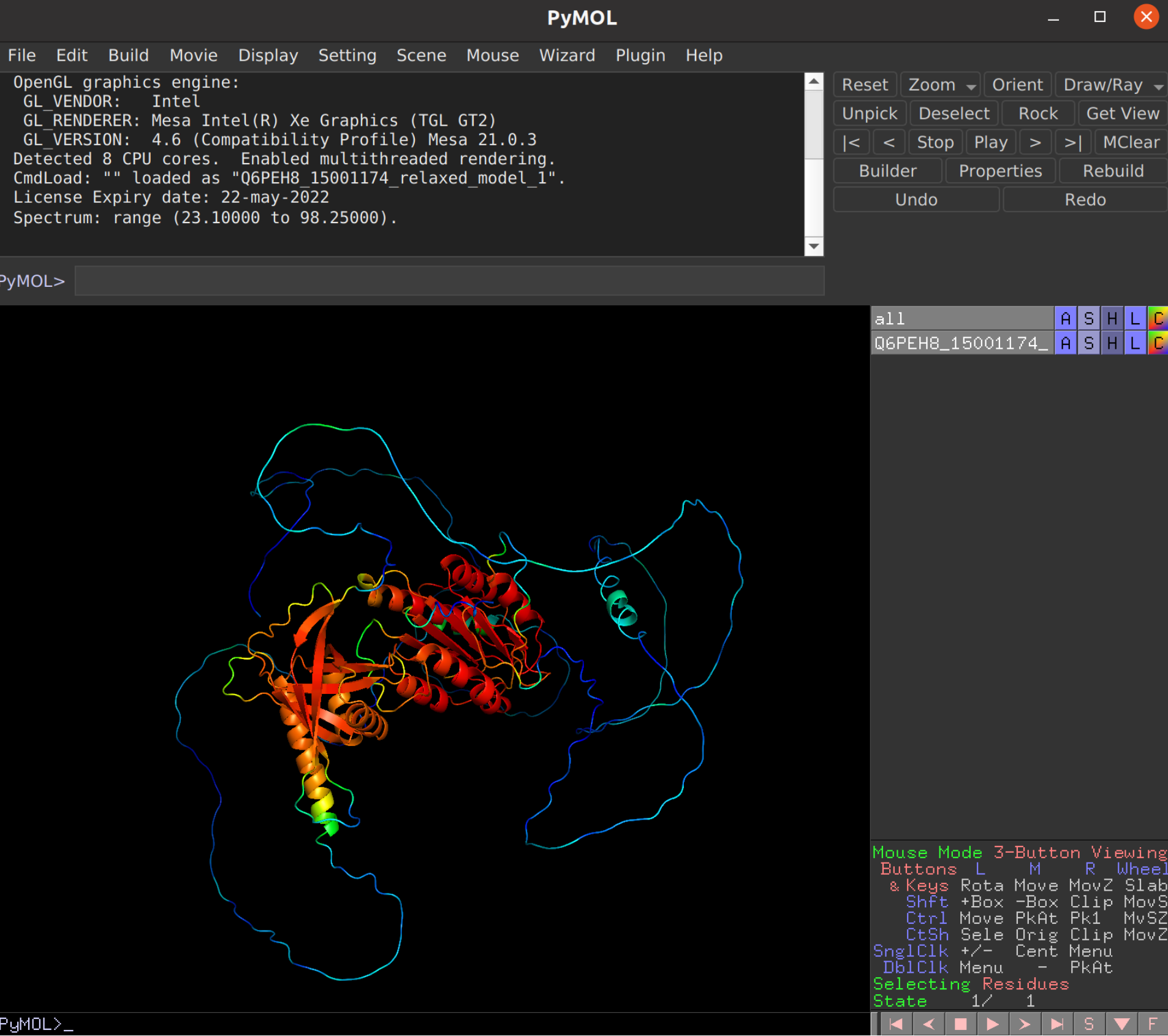
Alphafold Training Details E Learning Vib